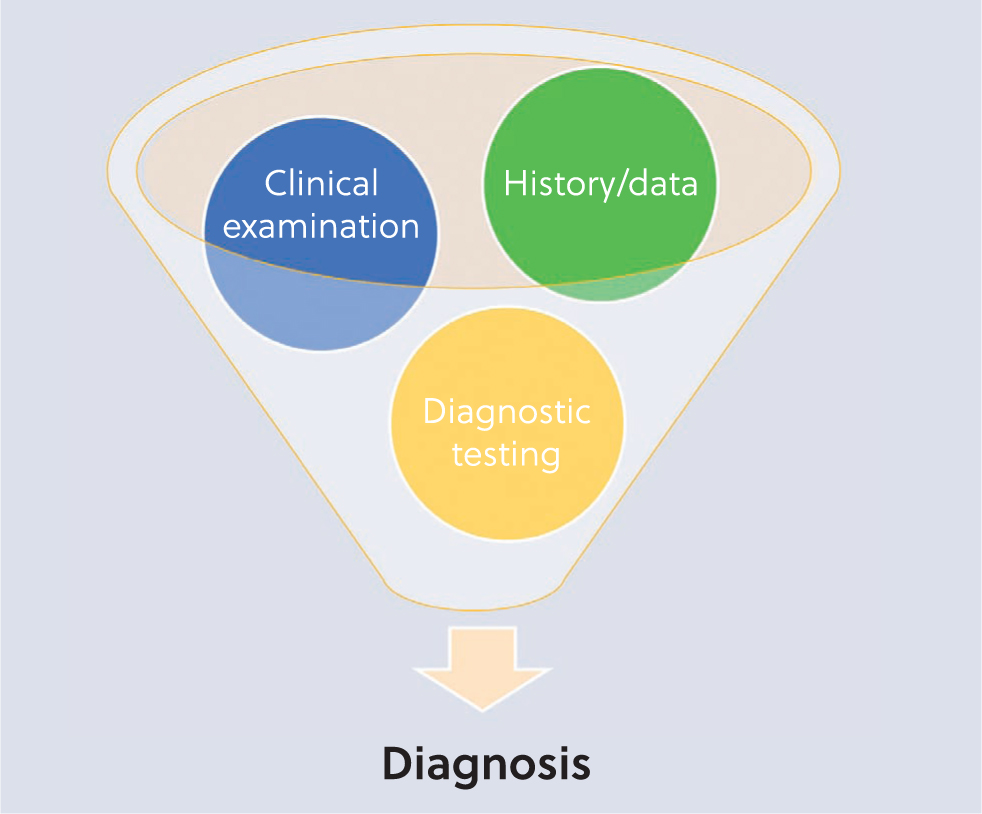
Disease diagnosis is a complex process, similar to hypothesis testing. Diagnostic tests should be considered ancillary measures, meaning they should complement a thorough process which culminates with a diagnosis, and includes a thorough history taking and/or proper data collection combined with, where necessary, clinical examination (Figure 1). These underlying steps will allow the clinician to formulate a comprehensive, yet relevant, list of differential diagnoses; diagnostic testing is then applied in a systematic, logical method to assess the list of differential diagnoses and confirm or exclude each possibility. Most importantly, diagnostic tests should not be used to stumble upon a diagnosis, but as a supportive tool in the decision-making process.
The fundamental, underlying question is, why do we need to perform diagnostic tests? The first reason will often be to confirm a clinical suspicion, either for personal interest or to apply the appropriate treatment and/or control measures. The classic example here is the development of anthelmintic resistance (Figure 2a); a very relevant topic in small ruminant medicine, where specific, updated guidelines are now available to guide diagnosis, providing reliable information for the selection of the appropriate anthelmintic class to use (Kaplan et al, 2023). The same applies in case of changes in routine management procedures or the application of preventive measures, where advice should be based on evidence of disease or on baseline measures to assess the impact of the proposed changes. For example, there are many vaccines available for small ruminants (Lacasta et al, 2015), therefore knowing which diseases are present on farm (or which have the highest impact), will be fundamental in selecting the most relevant vaccines to include in health management plans (Figure 2b). Finally, it is important to remember that, by submitting diagnostic samples to official laboratories (Figure 2c), clinicians can contribute to passive surveillance, providing data for monitoring of endemic diseases, as well as early detection of outbreaks and unknown diseases (Dijkstra et al, 2022).

Some key considerations to keep in mind when selecting the appropriate tests are:
- Cost-effectiveness
- The performance of the test(s)
- The sampling methodology.
Diagnostic challenges in small ruminant medicine
Selection of diagnostic tests
In small ruminant medicine, one of the main barriers to diagnostic testing is usually the cost of the test. At the time of writing, an individual test would cost from around £5 to almost £50, a price that does not take into account the cost of the vet or any other associated costs. Furthermore, the value of the animal(s) and the impact of the disease on the flock/herd will strongly influence the economic decision of testing and what test to use. Therefore, clinicians should be looking for the most cost-effective method of diagnosis, rather than the most economical. This would include not only the cost of the test itself, but also the repercussions of the disease on the overall economics of the farm (Nieuwhof and Bishop, 2005) and what strategies the clinician is planning to implement (e.g. control vs eradication).
The performance of the test(s), such as their sensitivity and specificity, their accuracy and the positive and negative predictive values, will also guide in deciding if that is the appropriate test for the situation. For example, a highly specific test will be most useful to rule in a disease, as it will have very few false positives, while a highly sensitive test will be needed to rule out or to demonstrate freedom from disease, as it will have very few false negatives (Cannon, 2001).
Finally, the sampling methodology should be considered, which includes questions like what biological material is needed, from which animals and the number of samples required. It is important to collect the most appropriate biological material, in the correct container and to ensure it is properly packaged (Table 1). A common example of incorrect selection and submission of samples to the laboratory is during abortion investigation (Borel et al, 2014), where submission of the placenta, along with other specimens, is crucial for reaching a diagnosis. However, to avoid collecting the wrong sample(s) or using the wrong container, it is always advisable to contact the laboratory to make sure the appropriate samples are collected and packaged (or at least check their brochure on the laboratory website). Another important consideration is whether to pool samples, which can provide a more costefficient solution in cases of parasitological examination of faecal samples (nematodes and liver flukes) or for flock/herd serological screening of infectious diseases. In this case, it is often best to collect individual samples and let the personnel at the laboratory pool the samples.
Table 1. Biological material commonly collected for diagnosis of small ruminants disease (in author's order of frequency), container used, special precautions and uses
| Biological material | Container | Special precaution | Most useful for |
|---|---|---|---|
| Serum | Plain vacutainer tube | Ideally 5 ml and stored in the fridge | Serology, biochemistry |
| Faeces | Plain plastic container | Minimum 5 g, freshly collected and stored in the fridge | Parasitology, microbiology |
| Urine | Plain plastic container | Biochemistry (glucose, protein, beta-hydroxybutyrate (BOHB)) | |
| Skin/hair | Plain plastic container | Parasitology | |
| Tissues | Plastic container with formalin | 1:10 tissue to formalin ratio and do not freeze | Histology |
| Tissues | Plain plastic container | Minimum 5 g and consider using transport medium or dried ice | Microbiology, biochemistry (cobalt, copper, selenium, lead) |
| Milk | Plain plastic container | Avoid contamination | Microbiology |
| Plasma | Heparin | Fill tube to the mark and mix well | Biochemistry (glutathione peroxidase, lead) |
| Whole blood | EDTA (ethylenediaminetetra-acetic acid) | Fill tube to the mark and mix well | Haematology |
| Vitreous humour | Plain vacutainer tube | Avoid blood contamination | Biochemistry (calcium, magnesium, BOHB, urea) |
Who to sample is obvious when dealing with a clinical suspicion in a single animal. However, for flock/herd investigations, sampling should be either based on targeted (or probability sampling) vs random (or non-probability) sampling. If we want to know the presence of a specific disease, for example, then targeted sampling would be the best option, where animals within the case definition are selected and sampled. The typical examples are so-called iceberg diseases, such as Johne's and lentivirus infection (Maedi visna and ovine pulmonary adenocarcinoma), which have similar syndromic signs (Busin, 2020). Clear, appropriate case definition – in the case in Figure 3, ill-thrift despite appropriate feeding and unremarkable clinical examination – becomes the important feature. If there are positive results and a highly specific test, it can be concluded that the disease is present; if there are negative results, however, freedom from the disease cannot be assumed.

However, to establish the extent (or the prevalence) of the disease or condition, then random sampling (where each individual has the same chances of being selected) is more appropriate. In the case of lentivirus infections, the extent of the disease within the herd/flock can be estimated (Kaba et al, 2023) and a decision taken on the most appropriate and cost-effective control measures.
Regarding the number of samples, it is common to use round numbers (e.g. 10 animals); however, statistical programmes for sample size calculations can also be used such as Epi Info (Centers for Disease Control and Prevention, 2023).
Available diagnostic tests
Compared to other animal species, especially companion animals or bovine, there might be either a reduced number of laboratories offering diagnostic tests for small ruminants or a limited number of tests commercially available (or validated in these species). In the UK, one of the laboratories within the national surveillance network (through the Animal Health and Plant Agency or Scotland's Rural College) can be used, or a laboratory associated with a veterinary school or a private diagnostic laboratory. They all have websites with brochures, usually with very detailed lists of tests offered, where and how to send samples, turn-around times and costs. Alternatively, many tests can be carried out in-house at the veterinary clinic or as ‘pen-side’ tests (Table 2). The use of point-of-care (POC) testing, which is a fast-growing field in veterinary diagnostics, is particularly suited for situations where facilities and funds are limited (Busin et al, 2016). POC will have considerable advantages over laboratory-based or centralised testing, which usually involves laborious and expensive techniques, trained personnel and transport of samples to dedicated facilities. Especially in the case of small ruminants, this can translate into more affordable veterinary care, reduced handling of animals, leading to targeted treatments and rapid testing in more remote geographic areas.
Table 2. Currently available small ruminants diagnostic tests, either at the vet practice or on farm (pen-side)
| Test | Target | Where |
|---|---|---|
| Biochemistry | Total protein, albumin, globulin, calcium, magnesium, urea, creatinine, beta-hydroxybutyrate, gamma-glutamyl transferase | Vet practice |
| Blood smear | Infection | Vet practice |
| Boray | Liver fluke | Vet practice |
| Faecal egg count | Nematodes | Vet practice |
| Fine needle aspirate | Infection, neoplasia | Vet practice |
| FMDV lateral flow device* | Foot-and-mouth virus | Pen-side |
| Haematology | Red blood cell, white blood cell, platelets | Vet practice |
| Ketone bodies with handheld meter | Beta-hydroxybutyrate | Pen-side |
| Refractometer | Hypogammaglobulinemia | Pen-side |
| Skin scraping | Mange, lice | Vet practice |
| Urine dipstick | Glucose, beta-hydroxybutyrate | Pen-side |
There is a wide variety of diagnostic tests available in small ruminant medicine (Table 3), from serology for antibody detection (mainly enzyme-linked immunosorbent assay (ELISA)), molecular techniques (mainly polymerase chain reaction (PCR)), parasitology, microbiology, biochemistry, haematology and anatomic pathology. The most appropriate diagnostic test(s) for each case under investigation should be chosen.
Table 3. Diagnostic tests currently commercially available for common targets/diseases for small ruminants in the UK and what type of sample can generally be submitted
| Target/disease | Test | Sample |
|---|---|---|
| Bibersteinia trehalosi | Microbiology (culture, serotyping and antibiotic sensitivity) | Tissues |
| Border Disease | Serology, molecular biology (polymerase chain reaction (PCR)) | Blood, tissues (spleen) |
| Campylobacteriosis | Microbiology (culture) | Faeces, tissues |
| Caseous lymphadenitis (Corynebacterium pseudotuberculosis) | Serology | Blood |
| Clostridium chauvoei, C. novyi, C. septicum, C. sordellii | Microbiology (fluorescent antibody test) | Tissues |
| Clostridium perfringens toxins | Microbiology (enzyme-linked immunosorbent assay (ELISA)) | Intestinal contents |
| Cobalt (vitB12), copper, selenium (GSH-Px) | Biochemistry | Blood, tissue (liver) |
| Coccidiosis | Parasitology (oocyst count + speciation) | Faeces |
| Cryptosporidiosis | Parasitology (smear) | Faeces |
| Enzootic abortion of ewes (Chlamydia abortus) | Serology, microbiology, molecular biology (PCR) | Blood, tissue (placenta) |
| Erysipelas (Erysipelothrix rhusiopathiae) | Serology (serum agglutination test) | Blood |
| Escherichia coli | Microbiology (culture) | Faeces, blood, milk |
| Footrot (Dichelobacter nodosus) | Molecular biology (PCR) | Swab |
| Giardiasis | Parasitology | Faeces |
| Haemonchus contortus | Parasitology (worm egg count + peptide nucleic acid test) | Faeces |
| Johne's (Mycobacterium avium subspecies paratuberculosis) | Serology (ELISA), molecular biology (PCR), microbiology (culture) | Blood, faeces |
| Lead | Biochemistry | Blood, tissue (liver/kidney) |
| Lice | Parasitology | Hair |
| Liver fluke (Fasciola hepatica) | Parasitology (Boray, liver fluke coproantigen ELISA), serology | Faeces, blood |
| Louping-ill | Serology (haemagglutination inhibition test) | Blood |
| Maedi visna (MV)/caprine arthritis encephalitis (CAE) | Serology (ELISA) | Blood |
| Mannheimia haemolytica/Pasteurella multocida | Microbiology (culture, serotyping and antibiotic sensitivity) | Milk, tissues |
| Mycoplasma conjunctivae | Molecular biology (PCR) | Swab |
| Mycoplasma ovipneumoniae | Microbiology, molecular biology (PCR) | Swab, tissues (lung) |
| Nematodes | Parasitology (worm egg count +/- larval speciation) | Faeces |
| ORF | Molecular biology | Tissue (scab) |
| Q fever (Coxiella burnetii) | Serology, molecular biology (PCR) | Blood |
| Red blood cell, haemoglobin, hematocrit, mean corpuscular volume, mean corpuscular haemoglobin, mean corpuscular hemoglobin concentration, red cell distribution width, white blood cell, platelet | Haematology | Blood |
| Salmonellosis | Microbiology (culture and serotyping) | Faeces, swabs, tissues |
| Schmallenberg (SBV) | Serology, molecular biology (PCR) | Blood, tissues (brain and spinal cord) |
| Sheep scab (Psoroptes ovis) | Serology (ELISA), parasitology | Blood, skin |
| Staphylococcus aureus | Microbiology (culture, serotyping and antibiotic sensitivity) | Swab, milk, tissues |
| Streptococcus dysgalactiae | Microbiology (culture, serotyping and antibiotic sensitivity) | Swab |
| Tick-borne fever (Anaplasma phagocytophilum) | Serology, molecular biology | Blood, tissue (spleen/lymph) |
| Total protein, albumin, globulin, calcium, magnesium, urea, creatinine, beta-hydroxybutyrate, gamma-glutamyl transferase, pepsinogen, zinc sulphate turbidity | Biochemistry | Blood |
| Toxoplasmosis (Toxoplasma gondii) | Serology, molecular biology (PCR) | Blood, tissue (brain) |
Serology is one of the most used, which tends to be rapid, low-cost and, usually, has good sensitivity and specificity. One of the major drawbacks is the persistence of antibodies over time (despite successful treatment or resolution of the disease) or the lack of differentiation between vaccinated and naturally infected animals. A blood sample in a plain vacutainer (red top) tube is usually needed. Common serological tests requested in everyday practice include those for:
- Enzootic abortion of ewes
- Toxoplasmosis
- Sheep scab
- Johne's
- Maedi visna (MV)/caprine arthritis encephalitis
- Caseous lymphadenitis
- Border disease.
Parasitology is probably comparable in frequency, as most clinicians will have either carried out or requested a faecal egg count (FEC) using a McMaster and microscope, as well as a Boray technique to identify liver fluke eggs or a skin scraping test for sheep scab/lice. Another common method is microbiology, where submitted samples (usually faeces, milk or blood) will be cultured to identify bacteria and advise on antibiotic treatment. A major limitation, in this case, would be any prior antibiotic treatment, which might prevent culturing of the bacteria.
Clinicians can also rely on biochemistry and haematology. These are not used as extensively as in small animal practice, but can provide useful information. Commonly used tests include albumin (ALB), beta-hydroxybutyrate (BHOB), urea, calcium (Ca) and magnesium (Mg) for ewes pre-lambing and trace elements deficiencies (cobalt (Co), copper (Cu) and selenium (Se)) for ill-thrift in growing animals. Other biochemistry parameters and haematology are usually only used when dealing with a single, valuable animal.
In cases of flock investigation, it is worth noting that packages are available, which are usually cheaper than ordering individual tests and these can guide the clinician in what are the appropriate tests for a syndromic condition (eg ewes metabolic profile, illthrift profile, enteric diseases).
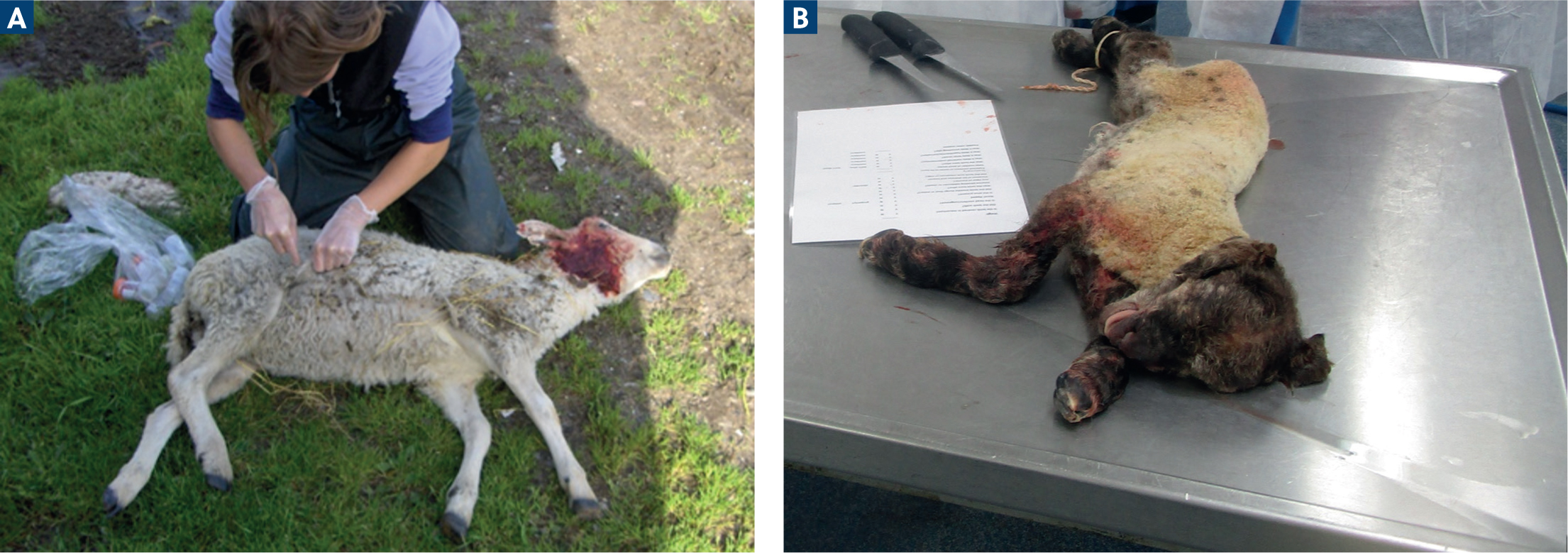
One final, important mention for small ruminants is the value of post-mortem investigations, either on farm (Figure 4a) or at diagnostic laboratories (Figure 4b). These are very cost-effective methods, which allow for many diseases to be checked at once, for the collection of multiple samples and they are often subsided by the government within the surveillance system. In this case, it is crucial to select the appropriate animals to sample and avoid any delay between death and post-mortem examination.
The future of diagnostics
The field of diagnostics, like many others, is fast evolving and many exciting new technologies have arisen in the last few years, which will help to overcome some of the challenges within small ruminant diagnostics.
Matrix assisted laser desorption ionization-time of flight mass spectrometry (MALDI-ToF MS) is a technology that allows amino acid sequencing and it is used for the identification of microorganisms, such as bacteria or fungi. A portion of a colony of the microbe in question is placed onto a MALDI-ToF plate, which means that, currently, culture of the microorganism (in broth or agar plate) is still necessary. This is then overlaid with a matrix, fired with a laser and the mass spectra are analysed by a dedicated software and compared with a database for species determination (biotyping), with results usually available within minutes. This avoids the need for other more time-consuming and laborious identification methods (usually biochemical and molecular), as well as providing the possibility to identify less common (or unknown) pathogens. Although most of the databases available are derived from human medicine, MALDI-ToF could be applied to bacteria commonly encountered in small ruminant medicine (Randall et al, 2015).
Similar technologies – mass spectrometers – are also used in the field of proteomics. The proteome is the set of all proteins and their expression in a system (cell, tissue or an organism). As not all proteins are expressed at all times, but are dependent on physiological and environmental factors, proteomics can provide an excellent overall view of disease processes at the protein level. Alterations in the proteome of body tissues or fluids (eg serum or faeces) can be measured directly, so changes that occur in a disease state can be accurately pinpointed. This approach is a very powerful tool for early-stage diagnosis of disease (eg identification of markers of inflammation) or for the identification of novel diagnostic antigens, by screening serum from infected and uninfected individuals (Ceciliani et al, 2014).
Another area that has attracted attention is the use of isothermal amplification technologies, where the thermocycling step necessary in conventional PCR is eliminated, enabling faster reaction times and removing one of the most significant impediments to developing POC molecular diagnostics. In particular, loop-mediated isothermal amplification (LAMP) is possibly one of the most known and widely researched isothermal amplification methods. This technique has already been applied for Haemonchus contortus detection (Costa-Junior et al, 2022), one of the most important small ruminant parasites, and has also been coupled with a lateral flow platform (Zhang et al, 2019) or so called lab-on-a-chip devices to make these diagnostic tests available at the POC (Wang et al, 2020).
Alongside these advances in the realm of molecular diagnosis, next generation sequencing (NGS) platforms allow the generation and handling of an enormous amount of data at the molecular level (RNA and DNA sequencing), providing an unprecedented level of information to simultaneously detect mixed infections, identify mutations or novel pathogens. An example of the potential of these technologies is the Oxford Nanopore Technologies MinION sequencer, a portable sequencer that allows for real-time whole genome sequencing in the field. Although some constrains still remain, like error rates and the need for expert bioinformatic skills, the applications of NGS in the control of important transboundary diseases, like Peste des petits ruminants and foot-and-mouth, is already promising (Liu et al, 2021; Bold et al, 2023).
The potential of sensor technologies in providing population-based, tailored diagnostics can be summarised with the commonly used term precision livestock farming (PLF). Ruminal boluses for electronic identification (Hentz et al, 2014), accelerometers to detect changes in behaviours (Alvarenga et al, 2016), GPS measurements to prevent pregnancy toxaemia (Donovan et al, 2013) and radar sensors to diagnose lameness (Busin et al, 2019) are all examples of very promising applications worth watching out for.
Finally, artificial intelligence (AI) is probably the hottest topic of the moment (Loeb, 2023). Will artificial intelligence replace vets? Or can vets harness the potential of machine learning tools, such as those algorithms used in image analysis or the use of Chat Generative Pre-trained Transformer (ChatGPT), to support diagnostic testing in small ruminant medicine?
Conclusions
The field of veterinary diagnostic testing can be complicated to navigate, but it is fascinating, extremely useful and fast evolving. A systematic approach to disease diagnostics should be applied, no matter what animal species we are dealing with. However, in small ruminants, some key differences, notably the animal's economic value and the farming system, should be kept in mind when selecting the most appropriate tests to reach a diagnosis.
KEY POINTS
- Disease diagnosis is a complex, systematic process involving history taking and/or data collection, clinical examination and diagnostic sampling.
- When selecting the appropriate diagnostic test(s), some key considerations are the cost-effectiveness, the performance of the test(s) and the sampling methodology.
- Serology, parasitology, microbiology and post-mortem investigations are commonly used and very useful diagnostic tests in small ruminant medicine.
- Point-of-care testing (either at the veterinary practice or as ‘penside’) can be a very valuable alternative to some laboratory-based tests.
- The field of diagnostics is fast evolving and many new technologies have been (or can be) applied to small ruminant diagnostic testing.


